Clonal Interference in Heterogeneous Networks
Understanding how network structures interact with the evolution of competing lineages in clonal populations.
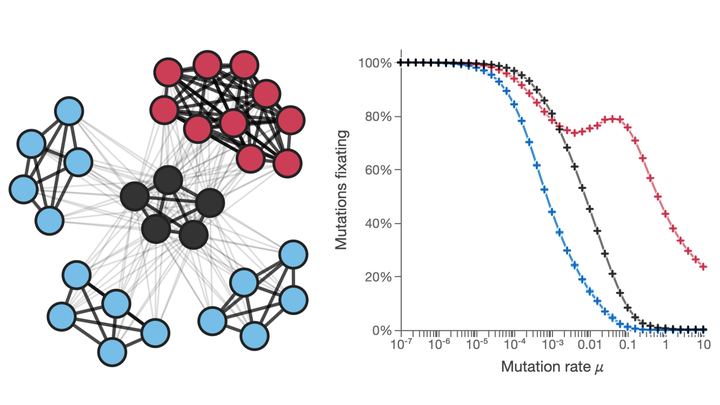
Our research group explores the intricate relationship between complex environmental structures and evolutionary processes, focusing on clonal interference in asexual populations. We aim to understand how organisms adapt in heterogeneous, network-like environments that closely resemble real-world scenarios.
Clonal interference (CI) occurs in asexual populations when multiple beneficial mutations arise and compete, unable to combine through recombination. While CI has been studied in well-mixed or simple spatial structures, its behavior in complex, heterogeneous environments remains largely unexplored.

Key Research Aims:
- Understanding CI in Highly Heterogeneous Environments: We investigate how diverse network topologies influence clonal interference.
- Identifying Key Structural Components: We aim to pinpoint specific elements of network structures that significantly impact adaptive processes, understanding which features promote faster adaptation and which areas are likely sources of adaptive mutations.
- Shaping and Controlling Adaptive Evolution: By understanding how network structures influence evolution, we seek to develop strategies for potentially guiding adaptive processes in complex environments.
- Developing Analytical Tools: We’re working to create and refine mathematical and computational tools that effectively model and predict evolutionary dynamics in intricate systems, bridging network theory, evolutionary biology, and complex systems science.
The insights gained from our research have potential applications across a wide range of fields. In epidemiology, our understanding of how pathogens evolve in structured host populations could form more effective disease management strategies. Conservation biologists might use our findings to guide habitat management practices, supporting species adaptation and resilience in fragmented environments. Our work could also lead to improved bioprocess designs by accounting for how environmental structure affects microbial evolution.
Beyond these direct applications, the principles we uncover might have broader, more speculative implications. For instance, our approaches to mutation spread in biological networks could provide insights into how content goes viral on social media platforms with diverse user structures. The competition between genetic strains in our research might elucidate how ideas, innovations and cultural practices propagate through social and professional networks.
As we continue to explore the intricate interplay between network complexity and evolutionary dynamics, we’re not merely advancing theoretical knowledge. We’re developing a deeper, more nuanced understanding of how adaptation occurs in complex systems. This research has the potential to open new avenues for managing and harnessing evolutionary processes in various fields, spanning from biology to the social sciences. By bridging the gap between network theory and evolutionary biology, we’re paving the way for a more comprehensive understanding of how information – be it genetic, cultural, or technological – spreads and competes in our interconnected world.